R package ggfastman
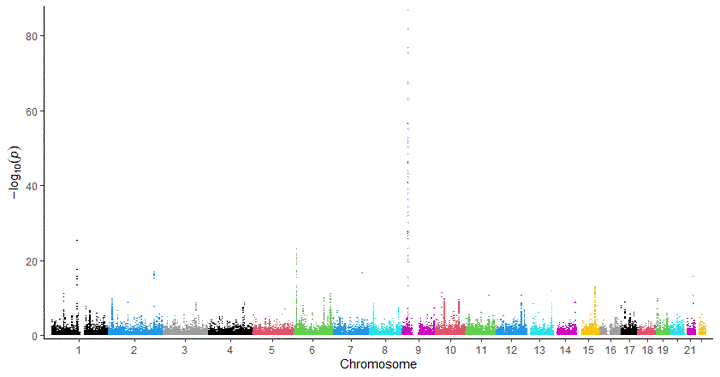
This is a very fast and easy-to-individualize plotting function for GWAS results e.g. pvalues. Since I’m using ggplot2 a lot, I adopted the idea from a very nice project from boxiangliu and combined it with a super fast plotting approach from the scattermore project.
A manhattan plot displays pvalues chromosomal positions against -mostly -log10 values- of genome-wide association studies between single nucleotide variants (SNV) or polymorphisms (SNP) and an endpoint e.g. expression, enzyme activity or case-control data.
One of the first R packages offering manhattan as well as qq plots was qqman from Stephen Turner, and nowadays there are a lot of different packages and approaches available for R and python. But a very fast one, which is still fast when plotting billions of data points, is still missing.
This package ggfastman is trying to fill this gap.